Undergraduate Minor
The Institute for Computational Medicine is proud to offer an Undergraduate Minor in Computational Medicine, the first educational program in CM, reflecting Johns Hopkins University’s leadership in this field. Like the ICM itself, the Undergraduate Minor in Computational Medicine is integrative and multidisciplinary. The ICM Core Faculty who serve as advisors to the Undergraduate Minor in Computational Medicine hold primary and joint appointments in multiple Johns Hopkins University departments and schools including Biomedical Engineering, Computer Science, Electrical and Computer Engineering, Mechanical Engineering, Applied Mathematics and Statistics (WSE); Neurosurgery, Emergency Medicine, Medicine, and the Divisions of Cardiology and Health Sciences Informatics (SOM); and Health Policy and Management (BSPH).
Undergraduates who are interested in learning about the Minor in Computational Medicine are encouraged to attend the ICM’s annual Computational Medicine Night.
Undergraduates who would like to begin the minor declaration process should complete the CM Minor Prospective Students Form.
Click here to listen to a Question and Answer session regarding the minor.
Click here to view a PDF of a sample curricula by major.
What will I gain from a minor in CM?
With a minor in CM, students will have a solid grounding in the development and application of computational methods in multiple key areas of medicine. Specifically, they will understand how mathematical models can be constructed from biophysical laws or experimental data, and how predictions from these models facilitate diagnosis and treatment of a disease. Graduating students will be conversant with a wide variety of statistical, deterministic and stochastic modeling methods. They will be able to develop a model and to write code to implement it; they will be able to analyze and visualize the resulting data from the simulations. These skills are essential to the advancement of modern medicine, and are prized both in academic research and industrial research. The courses and research opportunities available in the CM minor will place students at the forefront of the application of mathematics, computing and engineering to human health. Whether you go on to medical school, graduate research, or biomedical industries, the comprehensive quantitative training and exposure to cutting edge CM techniques will give you a competitive advantage for working in the medicine of tomorrow – which will be data-driven, predictive, personalized and preventative.
Will I have opportunities to specialize within CM?
Yes. The minor will provide both foundational training and opportunities for specialization in Computational Medicine. Students can select electives from an approved list that match their interests. We also provide examples of curricula in some key subareas of Computational Medicine, including:
Computational Physiological Medicine develops mechanistic models of biological systems in disease, and applies the insights gained from these models to develop improved diagnostics and therapies. Therapies could be diverse drugs, electrical stimulation, mechanical support devices and more.
Computational Molecular Medicine harnesses the enormous amount of disease-relevant data produced by next-generation sequencing, microarray and proteomic experiments of large patient cohorts, using statistical models to identify the drivers of disease and the susceptible links in disease networks.
Computational Anatomical Medicine uses medical imaging to analyze the variation in structure of human organs in health and disease. Such image analysis has been integrated into clinical workflows to assist in the diagnosis and prognosis of complex diseases.
Computational Healthcare is an emerging field devoted to understanding populations of patients and their interaction with all aspects of the healthcare process.
Techniques for and applications in each of these four key subareas will be introduced in the required core courses, so that students will be exposed to the breadth of Computational Medicine, and will be able to identify preferred areas of interest.
Who can declare the computational medicine minor?
The minor is available to all Whiting School of Engineering (WSE), Krieger School of Arts and Sciences (KSAS), and School of Medicine (SOM) premedical undergraduates. Students should have sufficient mathematical and programming background and should plan to complete the minor prerequisites during their first two years of study in order to be prepared for the minor.
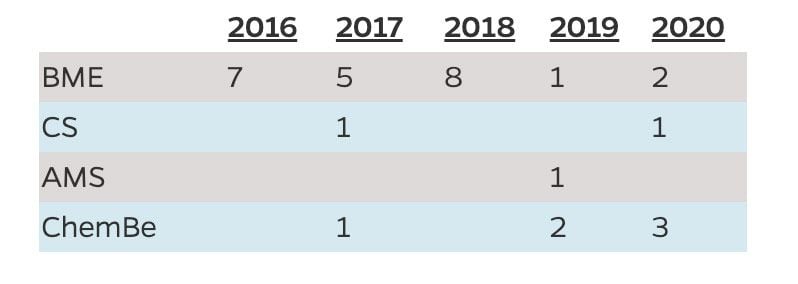
Number of Students Who Graduated with the Minor by Department
What are the prerequisites for the minor?
Before attempting the minor, undergraduates will have taken the following courses. For a course to count towards the minor, a minimum grade of C- is required. (Note: In general, courses graded as ‘S/U’ do not satisfy prerequisites, however exceptions will be made for courses taken during semesters in which all courses were graded S/U due to pandemic restrictions):
- Calculus I
- Calculus II
- Intermediate Probability and Statistics: either a single course covering both (EN.553.311, EN.560.348, or EN.540.382) or a course devoted to each (EN.553.420 and EN.553.430).
- At least one additional math or applied mathematics course (at least 3 credits)
- At least one of the following biological sciences course at the 200 level or higher (at least 3 credits):
| Biochemistry & Molecular Engineering (EN.580.221) |
| Biochemistry (AS.020.305) |
| Biochemistry I (AS.250.315/ AS.030.315) |
| The Nervous System (AS.080.305) |
| Biological Physics (AS.171.310) |
| Protein Engineering and Biochemistry Lab (AS.250.253) |
| Protein Biochemistry and Engineering Laboratory (AS.250.254) |
| Genetics (AS.020.303) |
6. At least one of the following computer programming course (at least 3 credits):
| Gateway Computing (EN.500.112, 113 or 114) |
| Computation & Programming for Materials Science & Engineering (EN.510.202) |
| Scientific Computing with Python (EN.553.383) |
| Scientific Computing: Linear Algebra (EN.553.385) |
| Scientific Computing: Differential Equations (EN.553.386) |
| Introduction to Scientific Computing for BME in Python, Matlab & R (EN.580.200) |
| Intermediate Programming (EN.601.120) |
| Intermediate Programming in Java (EN.601.107) |
| Data Structures (EN.601.226) |
What are the core classes needed for the minor?
1. Both of the following are required core courses for the minor and are usually completed junior or senior year:
| Course Num. | Course Name | Semester | Credits |
| EN.580.431 | Introduction to Computational Medicine: Imaging | 1st half of Fall | 2 |
| EN.580.433 | Introduction to Computational Medicine: The Physiome | 2nd half of Fall | 2 |
2. In addition to the courses listed above, one of the following is required:
| Course Num. | Course Name | Semester | Credits |
| AS.110.445 | Mathematical and Computational Foundations of Data Science | Spring | 4 |
| EN.553.450 | Computational Molecular Medicine | Spring | 4 |
| EN.580.430 | Systems Pharmacology & Personalized Medicine | Spring | 4 |
| EN.580.447 | Computational Stem Cell Biology | Spring | 3 |
| EN.580.458 | Computing the Transcriptome | Spring | 3 |
| EN.580.488 | Foundations of Computational Biology and Bioinformatics | Spring | 4 |
| PH.140.628 (71) & PH.140.629 (01) |
Data Science for Public Health I & II | Spring | 4 |
Am I required to attend ICM seminars?
Distinguished Seminar Series
In addition to the elective requirements, students with a declared Computational Medicine minor are REQUIRED to attend no less than 6 ICM Distinguished Seminars in person prior to graduation. Documentation of seminar attendance is two-fold: (1) Students must sign-in at every seminar attended and (2) students must complete the online Seminar Attendance Form. Please note that undergraduates do not need to register for the Distinguished Seminar Series in Computational Medicine course (EN.580.736/7) but do need to attend six ICM seminars and document their attendance to graduate with a Computational Medicine minor.
For the Fall 2021 semester, the seminar attendance requirement has been updated as follows:
ICM seminars will continue to be presented remotely via Zoom this semester. CM minors will be expected to ‘attend’ the live seminars scheduled for the Fall 2021 semester via Zoom. For students in time zones where viewing the seminar live is not feasible, the Fall 2021 seminars can be viewed as recorded. For all Fall 2021 seminars viewed, students must complete a seminar attendance form to receive credit towards the computational medicine minor.
More information on seminar speakers, dates, and topics can be found here.
What are the elective requirements for the minor?
Specific questions regarding the minor can be directed to icm@jhu.edu.


