Bio
“An Adaptable Framework to Extract Abnormal Brain Networks”
 Archana Venkataraman is an assistant professor in the Department of Electrical and Computer Engineering at Johns Hopkins University. She directs the Neural Systems Analysis Laboratory and is affiliated with the Malone Center for Engineering in Healthcare. Her research lies at the intersection of multimodal integration, network modeling and clinical neuroscience. Her objective is to develop a comprehensive and system-level understanding of the brain by strategically combining analytical tools, such as matrix factorization, signal processing and probabilistic inference, with application-driven hypotheses. This approach promises to yield novel insights into debilitating neurological disorders, with the long-term goal of improving patient care. Archana completed her B.S., M.Eng. and Ph.D. in Electrical Engineering at MIT in 2006, 2007 and 2012, respectively. She is a recipient of the CHDI Grant on network models for Huntington’s Disease, the MIT Lincoln Lab campus collaboration award, the NIH Advanced Multimodal Neuroimaging Training Grant, the National Defense Science and Engineering Graduate Fellowship, the Siebel Scholarship and the MIT Provost Presidential Fellowship.
Archana Venkataraman is an assistant professor in the Department of Electrical and Computer Engineering at Johns Hopkins University. She directs the Neural Systems Analysis Laboratory and is affiliated with the Malone Center for Engineering in Healthcare. Her research lies at the intersection of multimodal integration, network modeling and clinical neuroscience. Her objective is to develop a comprehensive and system-level understanding of the brain by strategically combining analytical tools, such as matrix factorization, signal processing and probabilistic inference, with application-driven hypotheses. This approach promises to yield novel insights into debilitating neurological disorders, with the long-term goal of improving patient care. Archana completed her B.S., M.Eng. and Ph.D. in Electrical Engineering at MIT in 2006, 2007 and 2012, respectively. She is a recipient of the CHDI Grant on network models for Huntington’s Disease, the MIT Lincoln Lab campus collaboration award, the NIH Advanced Multimodal Neuroimaging Training Grant, the National Defense Science and Engineering Graduate Fellowship, the Siebel Scholarship and the MIT Provost Presidential Fellowship.
 Click here to view webcast.
Click here to view webcast.
Abstract
“An Adaptable Framework to Extract Abnormal Brain Networks”
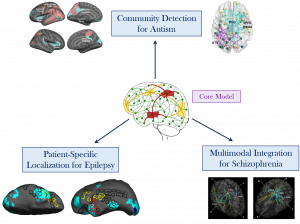
There is increasing evidence that complex neurological disorders reflect distributed impairments across multiple brain systems. These findings underscore the importance of network-based approaches for functional data. However, network analyses in clinical neuroimaging is largely limited to aggregate measures, which do not pinpoint a concrete etiological mechanism. In contrast, I will present a novel Bayesian framework that captures the underlying topology of the altered functional pathways.
In the first part of this talk, I will introduce our core framework to extract abnormal network foci from functional MRI data. This model relies on a latent structure, which captures hidden interactions within the brain; the latent variables are complemented by an intuitive likelihood model for the observed neuroimaging measures. The resulting variational EM algorithm produces clinically meaningful results by simultaneously localizing the centers of abnormal activity and the network of altered connectivity. Next, I will address three technical challenges: flexible network topology, multimodal integration and patient-specific analysis. I will demonstrate that our core framework can elegantly be adapted to each of these scenarios and yields novel insights into autism, schizophrenia and epilepsy, respectively. Finally, I will highlight some exciting future directions for our methodology that revolve around clinical understanding and interventions.
 Click here to view webcast.
Click here to view webcast.