Bio
“Computational Chemistry and Machine Learning in Molecular Medicine and Healthcare: Applications from the Molecule to the Patient”

Professor Collin M. Stultz is a principal investigator in the Research Laboratory of Electronics (RLE) at the Massachusetts Institute of Technology (MIT). He received his AB from Harvard College in 1988, his MD from Harvard Medical School in 1997, and his PhD in Biophysics with Martin Karplus at Harvard University, also in 1997. He is a board certified cardiologist who continues to see patients at Hospitals in the Boston area. Professor Stultz is on the faculty of MIT’s Department of Electrical Engineering and Computer Science (EECS) and the Institute for Medical Engineering and Science (IMES). He is a member of the American Society for Biochemistry and Molecular Biology and the Federation of American Societies for Experimental Biology. Among his honors are being a recipient of the Burroughs Welcome Fund Career Award in Biomedical Sciences, the NSF Career Award, and the Steven G. and Renée Finn Faculty Research Innovation Fellowship.
 Click here to view webcast.
Click here to view webcast.
Abstract
“Computational Chemistry and Machine Learning in Molecular Medicine and Healthcare: Applications from the Molecule to the Patient”
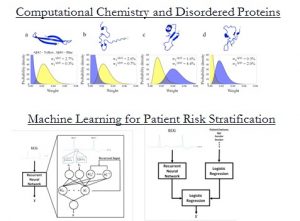
New computational methods coupled with increasing computing power are revolutionizing medicine and biomedical research. At the molecular scale, computational models allow us to probe fundamental biophysical processes at an unprecedented level of detail, garnering insights that would have been difficult to obtain from experiment alone. At the patient or population scale, computation (and machine learning in particular) provides a platform for learning models that can help physicians choose the best therapies for their patients. In this seminar I will attempt to illustrate these concepts using work from our own research group.
In the first half of this seminar I describe a new method, which is based on Bayesian statistics, for modeling the structure of flexible proteins like Aβ42 – an intrinsically disordered protein (IDP) that plays a role in the pathogenesis of Alzheimer’s disease. While disordered and monomeric in solution, Aβ42 forms neurotoxic soluble aggregates in the brains of patients with Alzheimer’s disease. Through a series of computational and experimental studies we deduced fundamental steps that are important in the aggregation process. These data provide a detailed mechanistic description of the amyloid-β aggregation pathway and suggest a structural link between the disordered free monomer and the growth of amyloid fibrils and soluble oligomers. In the second half of this seminar I demonstrate how new insights, which can be used to guide clinical decision-making, may be obtained with machine learning. We focus on the problem of identifying patients who are at high risk of adverse cardiovascular events after an acute coronary syndrome (ACS) – a clinical condition characterized by reduced blood flow to the heart. Using a database of over 5000 patients who have suffered an ACS, we developed a neural network model that is able to identify high-risk patient subgroups. Such models provide clinicians with a rich source of information that can be exploited to ensure that patients receive therapies that are appropriate for their level of risk.
 Click here to view webcast.
Click here to view webcast.