Bio
“Designing Intact Cardiac Cell Electrophysiological Protocols for the Development of more Robust Computational Models”
 Dr. David Christini received a BS degree in electrical engineering from the Pennsylvania State University and MS and PhD degrees in biomedical engineering from Boston University. He is a Professor in the Departments of Medicine and Physiology and Biophysics, Weill Cornell Medical College, New York. His group’s efforts are focused on improving the understanding of, and therapies for, cardiac arrhythmias. Their experiments employ electrophysiological and imaging modalities, primarily at the cellular and tissue levels. They combine experimental data with clinical data to inform computational cardiac modeling that scales up from the single cell to the whole organ. Such a combination of experimental, computational, and clinical approaches provides the synergy to impact understanding of cardiac arrhythmias. He is also Vice Dean of the Weill Cornell Graduate School. More information is available at christinilab.org.
Dr. David Christini received a BS degree in electrical engineering from the Pennsylvania State University and MS and PhD degrees in biomedical engineering from Boston University. He is a Professor in the Departments of Medicine and Physiology and Biophysics, Weill Cornell Medical College, New York. His group’s efforts are focused on improving the understanding of, and therapies for, cardiac arrhythmias. Their experiments employ electrophysiological and imaging modalities, primarily at the cellular and tissue levels. They combine experimental data with clinical data to inform computational cardiac modeling that scales up from the single cell to the whole organ. Such a combination of experimental, computational, and clinical approaches provides the synergy to impact understanding of cardiac arrhythmias. He is also Vice Dean of the Weill Cornell Graduate School. More information is available at christinilab.org.
 Click here to view webcast.
Click here to view webcast.
Abstract
“Designing Intact Cardiac Cell Electrophysiological Protocols for the Development of more Robust Computational Models”
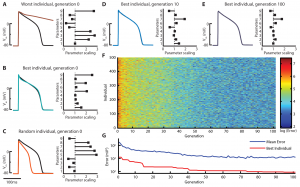
The traditional paradigm for developing cardiac computational cell models utilizes data from multiple cell types, species, laboratories, and experimental conditions to create a composite model. While such models can accurately represent data in limited biological scenarios, their ability to predict behavior outside of a narrow dynamic window is limited. This talk will describe the rationale behind using novel electrophysiological protocols that aim to densely sample the dynamics of intact cardiac myocytes. The information-rich data from such protocols are then fit using global parameter optimization algorithms to tune multiple model parameters simultaneously. By so doing, this approach yields cell models that fit wide-ranging cellular behavior, making them better suited for physiological and pathophysiological predictions.
 Click here to view webcast.
Click here to view webcast.