Bio
“Integrated Time-Course Omics Analysis Distinguishes Immediate Therapeutic Response from Acquired Resistance”
 Elana Fertig’s lab pursues research in the systems biology of cancer and therapeutic response. She develops computational methods for pattern detection from genomics data and integration of diverse high-throughput data modalities. These algorithms are applied to analyze data from diverse cancer types, with a primary focus on head and neck squamous cell carcinoma (HNSCC) and therapeutic resistance. Dr. Fertig is an Assistant Professor of Oncology at Johns Hopkins University, with affiliations in the Institute of Computational Medicine, Center for Computational Genomics, Machine Learning, and the Center for Computational Biology. Prior to entering the field of computational cancer biology, Dr Fertig was a NASA research fellow in numerical weather prediction. Dr. Fertig’s research is featured in numerous peer reviewed publications, R/Bioconductor packages, and independent research funding. She led the team that won the HPN-DREAM8 algorithm to predict phospho-proteomic trajectories from therapeutic response in cancer cells and her research has been featured in an article in US News and World Reports and Time Magazine.
Elana Fertig’s lab pursues research in the systems biology of cancer and therapeutic response. She develops computational methods for pattern detection from genomics data and integration of diverse high-throughput data modalities. These algorithms are applied to analyze data from diverse cancer types, with a primary focus on head and neck squamous cell carcinoma (HNSCC) and therapeutic resistance. Dr. Fertig is an Assistant Professor of Oncology at Johns Hopkins University, with affiliations in the Institute of Computational Medicine, Center for Computational Genomics, Machine Learning, and the Center for Computational Biology. Prior to entering the field of computational cancer biology, Dr Fertig was a NASA research fellow in numerical weather prediction. Dr. Fertig’s research is featured in numerous peer reviewed publications, R/Bioconductor packages, and independent research funding. She led the team that won the HPN-DREAM8 algorithm to predict phospho-proteomic trajectories from therapeutic response in cancer cells and her research has been featured in an article in US News and World Reports and Time Magazine.
 Click here to view webcast.
Click here to view webcast.
Abstract
“Integrated Time-Course Omics Analysis Distinguishes Immediate Therapeutic Response from Acquired Resistance”
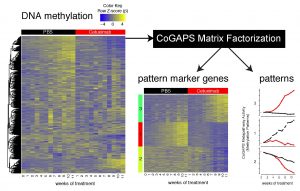 Targeted therapies specifically act by blocking the activity of genes that are critical for tumorigenesis. However, most cancers acquire resistance and long-term disease control is rarely observed. Knowing the timing of molecular changes responsible for the development of acquired resistance can enable optimization of alterations to patients’ treatments. Clinically, acquired therapeutic resistance can only be studied at a single time point in resistant tumors. To determine the dynamics of these molecular changes, we obtained high throughput omics data weekly during the development of cetuximab resistance in a head and neck cancer model. An unsupervised, Bayesian non-negative matrix factorization algorithm, CoGAPS, quantified the evolving transcriptional and epigenetic changes. Further applying a PatternMarker statistic to the results from CoGAPS enabled novel heatmap-based visualization of the dynamics in these time-course omics data. We demonstrate that transcriptional changes resulted from immediate therapeutic response and resistance whereas epigenetic alterations only occurred with resistance. Integrated analysis demonstrated delayed onset of changes in DNA methylation relative to transcription, suggesting that resistance was stabilized epigenetically. Conclusions: Genes with epigenetic alterations associated with resistance that had concordant expression changes were hypothesized to stabilize resistance. These genes include FGFR1, which was associated with EGFR inhibitor resistance previously. Thus, integrated omics analysis distinguishes the timing of molecular drivers of resistance. Our findings are a relevant step into the better understanding of the time course progression of changes resulting in acquired resistance to targeted therapies and for time course genomics analysis.
Targeted therapies specifically act by blocking the activity of genes that are critical for tumorigenesis. However, most cancers acquire resistance and long-term disease control is rarely observed. Knowing the timing of molecular changes responsible for the development of acquired resistance can enable optimization of alterations to patients’ treatments. Clinically, acquired therapeutic resistance can only be studied at a single time point in resistant tumors. To determine the dynamics of these molecular changes, we obtained high throughput omics data weekly during the development of cetuximab resistance in a head and neck cancer model. An unsupervised, Bayesian non-negative matrix factorization algorithm, CoGAPS, quantified the evolving transcriptional and epigenetic changes. Further applying a PatternMarker statistic to the results from CoGAPS enabled novel heatmap-based visualization of the dynamics in these time-course omics data. We demonstrate that transcriptional changes resulted from immediate therapeutic response and resistance whereas epigenetic alterations only occurred with resistance. Integrated analysis demonstrated delayed onset of changes in DNA methylation relative to transcription, suggesting that resistance was stabilized epigenetically. Conclusions: Genes with epigenetic alterations associated with resistance that had concordant expression changes were hypothesized to stabilize resistance. These genes include FGFR1, which was associated with EGFR inhibitor resistance previously. Thus, integrated omics analysis distinguishes the timing of molecular drivers of resistance. Our findings are a relevant step into the better understanding of the time course progression of changes resulting in acquired resistance to targeted therapies and for time course genomics analysis.
 Click here to view webcast.
Click here to view webcast.