Computational Molecular Medicine
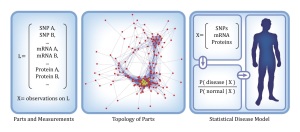 Understanding the function of highly interconnected molecular networks has come to be known as “systems biology.” Diseased cells arise from perturbations in biological networks owing to the net effect of interactions among multiple molecular agents, including inherited and somatic DNA variants; changes in mRNA, microRNA (miRNA), and protein expression; and epigenetic factors, such as DNA methylation. An enormous amount of data about these perturbations is being produced by next-generation sequencing and microarray experiments of large patient cohorts, making it possible for the first time to discover the driving differences in the abundance and activity of key biomolecules. Analysis of high-dimensional biomolecular data using methods of statistical learning has the potential to enhance discovery of molecular disease networks, detection of disease, discrimination among disease subtypes, prediction of clinical outcomes, and characterization of disease progression.
Understanding the function of highly interconnected molecular networks has come to be known as “systems biology.” Diseased cells arise from perturbations in biological networks owing to the net effect of interactions among multiple molecular agents, including inherited and somatic DNA variants; changes in mRNA, microRNA (miRNA), and protein expression; and epigenetic factors, such as DNA methylation. An enormous amount of data about these perturbations is being produced by next-generation sequencing and microarray experiments of large patient cohorts, making it possible for the first time to discover the driving differences in the abundance and activity of key biomolecules. Analysis of high-dimensional biomolecular data using methods of statistical learning has the potential to enhance discovery of molecular disease networks, detection of disease, discrimination among disease subtypes, prediction of clinical outcomes, and characterization of disease progression.
Owing to the massive number of interacting components in biological systems, the traditional approach to biomedical research – which is experimental and molecule by molecule – is not feasible for high-throughput assessment of biological complexity. A principled learning approach has become indispensable for extracting knowledge from large arrays of numbers. In the case of computational molecular medicine, this entails revealing and exploiting disease-related information implicitly stored in high-dimensional, high-throughput biological data.
A deep understanding also requires a statistical characterization: Learning the likely and unlikely concentrations of these bio-molecules – not just individually but collectively as a multivariate probability distribution – opens the possibility of making clinical decisions based on these likelihoods. Statistical modeling is motivated by the simple fact that in functioning organisms, not all combinations of individual molecular states are equally likely. Some configurations are observed far more likely that others, and thus, the favored states in health and disease are markedly different.


