Bio
“Pan-genomic Advances for Fighting Reference Bias”

Ben Langmead is an Assistant Professor of Computer Science at Johns Hopkins University. He earned a Ph.D. in Computer Science from the University of Maryland in 2012. His group seeks to make high-throughput biological datasets easy for biomedical researchers to use. The group studies and applies ideas from sequence alignment, text indexing, statistics and parallel programming. He has released several high-impact software tools (e.g. Bowtie, Bowtie 2) that address common genomics research questions. His paper describing Bowtie won the Genome Biology award for outstanding paper in 2009. He has also released scalable software tools (e.g. recount3, Snaptron) that use the MapReduce parallel programming model and commercial cloud computing services to analyze large collections of sequencing data. He is lucky to have excellent connections with many of the superb JHU BME faculty. He is the recipient of a Sloan Research Fellowship (2014), a National Science Foundation CAREER award (2014) and the Benjamin Franklin award for contributions to open access (2016).
▶RECORDING
Date/Time: Tuesday, September 3rd at 4:00PM EST
Location: Homewood Campus: Clark 110
Live Webcast: https://wse.zoom.us/j/97268201562
Abstract
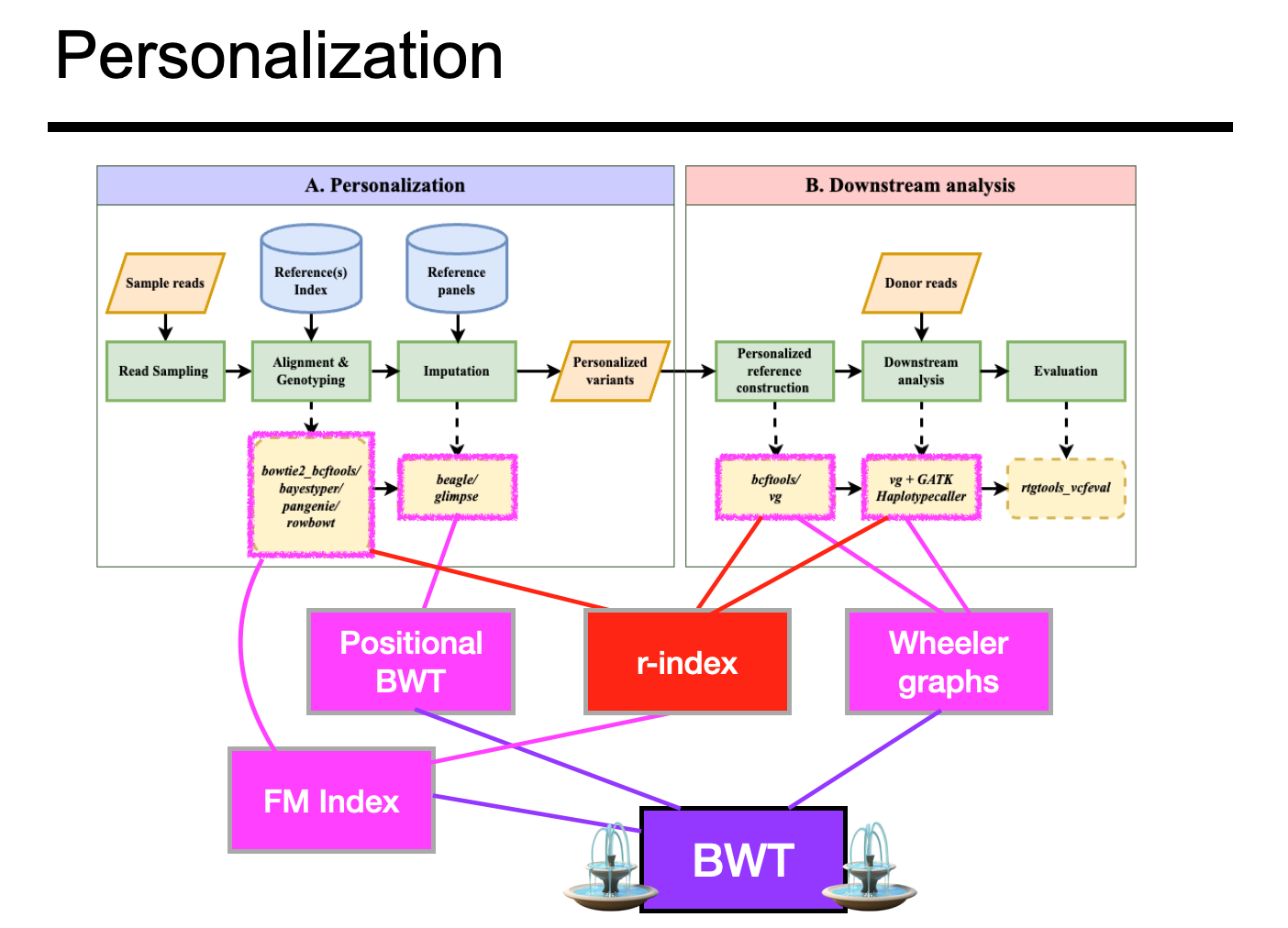
“Pan-genomic Advances for Fighting Reference Bias”
“Sequencing data analysis often begins with aligning reads to a reference genome, where the reference takes the form of a linear string of bases. But linearity leads to reference bias, a tendency to miss or misreport alignments containing non-reference alleles, which can confound downstream statistical and biological results. This is a major concern in human genomics; we don’t want to live in a world where diagnostics and therapeutics are differentially effective depending whether and where our genetic variants happen to match the reference. Fortunately, computer science and bioinformatics are meeting the moment. We can now index and align sequencing reads to references that include many population variants. I will present some of the major and insights that have shaped this journey from the early days of efficient genome indexing — especially the Burrows-Wheeler Transform — continuing through recent methods for indexing graph-shaped references and references that include many genomes. I will emphasize recent results that show how to optimize simple and complex pan-genome representations for effective avoidance of reference bias. Finally, I will outline promising methods for the bias, including new ideas for how to measure bias, new proposals in compressed indexing, and new workflows that integrate genotype imputation to improve reference bias.
Much of this work is collaborative with Travis Gagie, Christina Boucher, Alan Kuhnle and others.”
▶RECORDING
Date/Time: Tuesday, September 3rd at 4:00PM EST
Location: Homewood Campus: Clark 110
Live Webcast: https://wse.zoom.us/j/97268201562