Uncovering the Origins of Heterogeneity in Responses to MAPK-Targeted Cancer Therapies
Jump to:
Bio
“Uncovering the Origins of Heterogeneity in Responses to MAPK-Targeted Cancer Therapies”
Dr. Mohammad Fallahi-Sichani is a tenure-track Assistant Professor of Biomedical Engineering in the University of Virginia (UVa), Basic Science Lead of the Melanoma Research Team at UVA Cancer Center, and a member of the Molecular and Cellular Basis of Disease (MCBD) Graduate Program at UVa. Prior to relocating his laboratory to UVa in June 2020, he served as an Assistant Professor of Biomedical Engineering in the University of Michigan, beginning in Summer 2017. Since then, he has established a laboratory that currently comprises 1 research lab specialist, 3 PhD students, 1 MD/PhD student, 2 postdoctoral fellows, and 3 undergraduates. He received my Ph.D. in Chemical Engineering from the Univ. of Michigan in 2012. Under the joint supervision of Jennifer Linderman and Denise Kirschner, he developed multi-scale models of lung pathology to bridge molecular, cellular and tissue level mechanisms of immune response to M. tuberculosis infection. Dr. Fallahi-Sichani completed his postdoctoral training with Peter Sorger during 2012-2017, working as a Life Sciences Research Foundation (LSRF) Postdoctoral Fellow in Systems Biology at Harvard Medical School. He was then interested in understanding how responses of heterogeneous tumor cells to targeted therapeutics are determined at the level of their signaling networks. His postdoctoral training provided a great opportunity to expand his scientific expertise towards high-throughput and single-cell quantitative biology, cancer biology and pharmacology. Dr. Fallahi-Sichani developed new methods to combine live-cell imaging, multiplexed measurements, single-cell analysis, and data-driven computational modeling, to link the observed cell-to-cell heterogeneity in drug effect in tumors to the consequential changes in cell cycle regulation and acquisition of drug resistance phenotypes. Research in his own laboratory aims at designing, building, and utilizing new experimental and computational tools to discover fundamental mechanisms that regulate the behavior of human cells in response to perturbations, such as cytokines, environmental stress, and therapeutic drugs. By merging multiplexed, high-throughput, single-cell data acquisition with multi-scale computational modeling, he hopes to understand the molecular mechanisms that underlie adaptive cell fate decisions in the presence of cellautonomous, microenvironment, and therapy-induced selective pressures, and elucidate how they vary under unhealthy conditions, particularly in cancer cells. From a therapeutic point of view, a detailed understanding of these mechanisms will provide a rational basis for choosing the optimal therapeutic targets to: (i) maximize the desired effect in diseased cells (e.g., tumor cell killing), (ii) prevent the development of therapeutic resistance, and (iii) reduce therapy-induced adverse effects in healthy cells.
To join the live event please request the link by emailing: icm@jhu.edu
Abstract
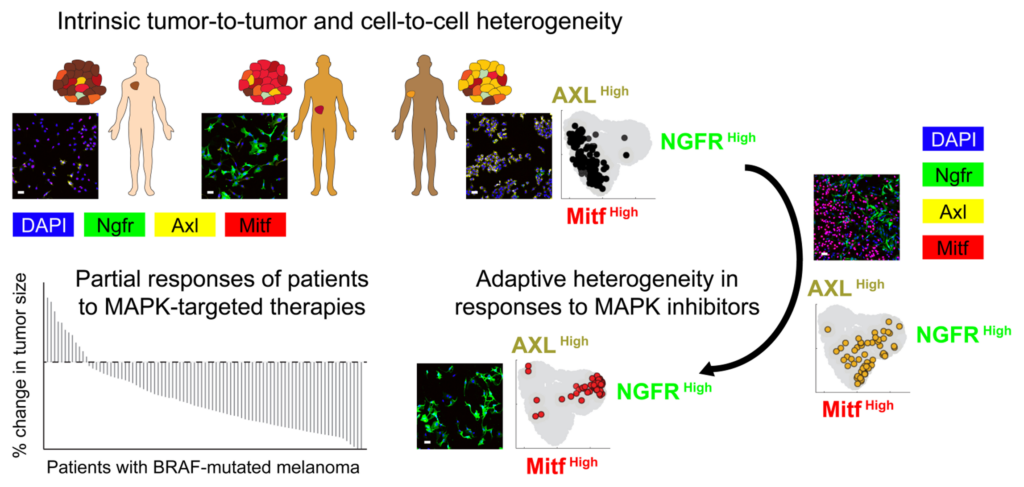
“Uncovering the Origins of Heterogeneity in Responses to MAPK-Targeted Cancer Therapies”
Tumor cells that carry the same mutated oncogenes respond nonidentically to therapeutic inhibitors of oncogenic signaling. This poses a major challenge to the use of targeted therapies, arguably the cornerstone of precision cancer medicine. Although genetic alterations may cause late drug resistance, emerging evidence suggests that tissue-specific, epigenetic mechanisms and rewiring of transcriptional networks rapidly induce drug-tolerant states with reduced dependency on the oncogenic activity. I will describe systems biology approaches to identify such states in the context of BRAF-mutated melanomas treated with BRAF/MAPK targeted therapies. By merging highly multiplexed single-cell assays, high-throughput data acquisition, and integrative computational modeling, we assemble an integrated picture that links molecular determinants of drug response to the diversity of intrinsic and adaptive phenotypes across both genetically diverse and isogenic populations of tumor cells. This picture will allow us to identify and test efficient ways to block subpopulations of drug-resistant cells, and thereby advance the development of more effective therapies.