Bio
“Quantitative Drug Discovery – Opportunities for Modeling and Simulation in the Age of Machine Learning”

Dr. Birgit Schoeberl currently serves a Global Head of Modeling and Simulation, PK Sciences at Novartis. Before joining Novartis, Birgit served as interim Senior Vice President, Scientific Value, at GNS Healthcare. During her time at GNS, she focused on applying causal machine learning to drug development. Prior to GNS, she was a member of the founding team at Merrimack Pharmaceuticals and where she spent over 12 years in different roles. In her latest role as Head of Discovery and Early Development, she gained broad experience in oncology drug discovery and development. Birgit and her leadership team were responsible for multiple preclinical and early clinical programs.
Birgit received her Chemical Engineering degree from the Technical University in Karlsruhe, Germany, and her Ph.D. in Biological Engineering from the Max Planck Institute for Dynamics of Complex Technical Systems in Magdeburg. She was a post-doctoral fellow in the laboratories of Douglas Lauffenburger and Peter Sorger in the Biological Engineering Department at MIT.
 Click here to view webcast.
Click here to view webcast.
Abstract
“Quantitative Drug Discovery – Opportunities for Modeling and Simulation in the Age of Machine Learning”
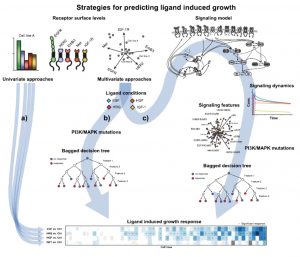
In my talk, I will discuss how different modeling approaches from machine learning to mechanistic computational models are applied throughout the drug discovery process. We discuss a proliferation screen across 58 cell lines stimulated with multiple ligands. At this example, we explore the potential to combine machine learning and mechanistic computational models. We show that the predictive model performance increases when simulated features derived from the mechanistic model are included as features into a Bagged Decision Tree (BDTs) model. The presented novel approach of using BDTs in conjunction with simulated signaling features is the beginning of how complex mechanistic models and large data sets can be combined to understand cellular complexity.
 Click here to view webcast.
Click here to view webcast.