Bio
“Mechanistic Modeling of Signal Transduction and Dynamic Cell Morphologies”

Dr. Meier-Schellersheim’s research group has been developing tools for quantitative computational image analysis and mechanistic modeling of cellular behavior for several years now. They were able to develop computational methods and tools that permit the simulation of cellular signaling networks embedded into dynamic, realistic 3D morphologies to take into account the coupling between cellular biochemistry and morphological dynamics. The simulation platform Simmune was the first modeling software to permit this degree of realism and we have continuously been improving its capabilities with regard to the size of multi-cellular systems that can be simulated and parameter scans that can be performed on distributed computer systems. Simmune can model cells that express receptors for chemosensing and adhesion on their surface that react to receptor mediated stimuli by adjusting their geometry to adhere to extracellular structures or by directed migration in response to chemotactic signals. Although the coupling between biochemistry and cell motion based on Potts model rules is phenomenological applying such simulation techniques to explore the role cell-cell and cell-matrix interactions may play in complex 3D structures is a first step towards understanding how chemical and mechanical cues regulate cellular migration. Dr. Meier-Schellersheim’s group develops and applies quantitative image analysis tools to extract the information needed for detailed spatially resolved simulations directly in an unbiased and automated manner from image data. Having been part of the Laboratory of Systems Biology for several years now, his group acquired considerable experience in coordinating interdisciplinary work and in handling large heterogeneous data sets.
In addition to exploring the interplay between cell morphology and cellular responses towards stimuli, Dr. Meier-Schellersheim’s group develops and applies tools that can perform systematic analyses of the behavior of computational models of cellular signaling pathways. These tools can identify which elements of a pathway model are responsible for reproducing specific features in the experimentally observed cellular behavior.
To join the live event please request the link by emailing: icm@jhu.edu
▶RECORDING
Abstract
“Mechanistic Modeling of Signal Transduction and Dynamic Cell Morphologies”
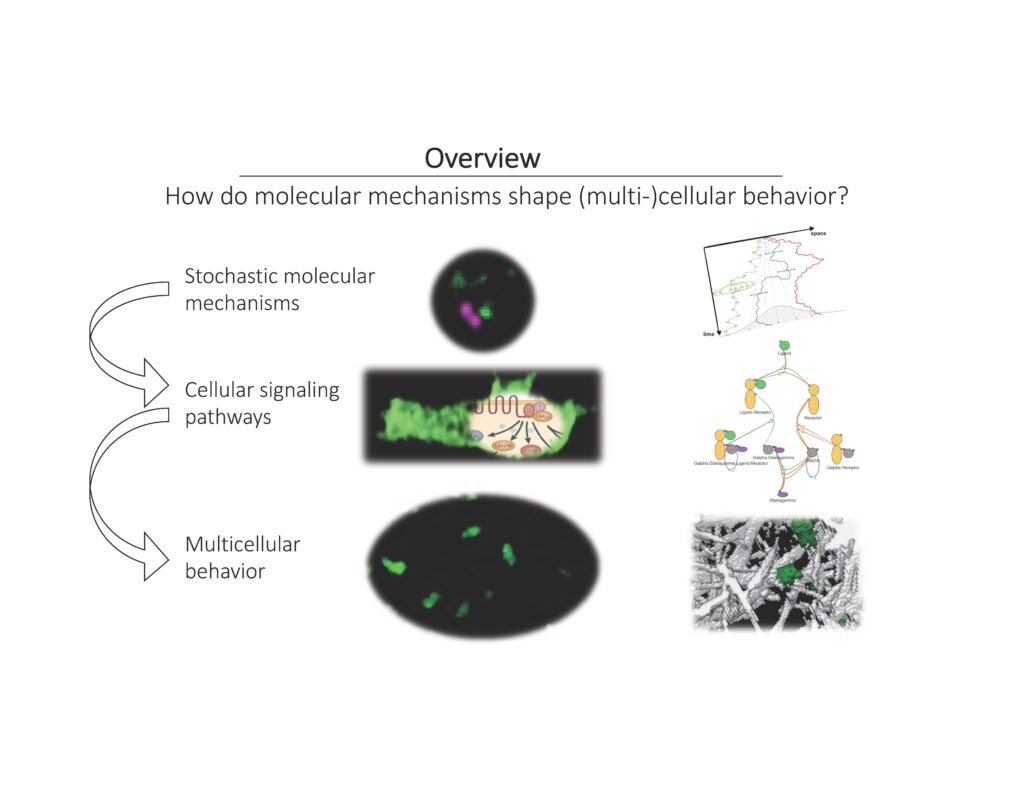
Detailed mechanistic models of cellular signaling pathways have the advantage that the conclusions they allow us to draw can be tested with little ambiguity. However, building such models typically requires more data and involves far more parameters than phenomenological approaches do. Using examples from cytokine and growth factor signaling, I will discuss some recent progress in applying detailed modeling tools and describe what can be learned from models whose parameters cannot be uniquely determined. Then, I will show how detailed models of intracellular biochemistry can be linked to a realistic treatment of morphological dynamics using a novel approach for representing cellular surfaces.
▶RECORDING