The Landscape of the Heritable Cancer Genome
email [email protected] for link
Jump to:
Bio
“The Landscape of the Heritable Cancer Genome”
Giovanni Stracquadanio is an UKRI EPSRC fellow, Senior Lecturer (Associate Professor) in Synthetic Biology and co-director of the Edinburgh Genome Foundry (EGF). He group is interested in understanding the molecular mechanisms underpinning complex phenotypes and diseases using two of the most disruptive technologies available: synthetic biology and machine learning. His long-term goal is to reverse-engineer biological systems to develop generative algorithms to design, build and test biological agents for addressing healthcare problems, such as rare metabolic diseases and cancer, and industrial biotechnology challenges, like de-novo enzyme engineering.
Dr Stracquadanio obtained a PhD in Informatics from the University of Catania (Italy) in 2010, working on global optimisation methods for protein structure prediction and metabolic engineering. He then received postdoctoral training in synthetic biology in Joel Bader and Jef Boeke labs at the Johns Hopkins University working on the synthetic yeast genome.
Dr Stracquadanio was a main contributor to the Synthetic Yeast (Sc2.0) genome project, pioneering algorithms and developing software at the foundation of the first synthetic eukaryotic genome. He has also developed tools used in large-scale synthesis projects, streamlining chromosomes engineering and the assembly of biological pathways.
In 2014, he moved to the Ludwig Institute for Cancer Research at the University of Oxford UK to work on cancer genetics, in Gareth Bond’s lab; here, he focused on studying how high-frequency inherited p53 mutations affect the risk of cancer and response to treatment using statistical genetics methods and cell line models.
In 2016, prior to joining the University of Edinburgh, Dr Stracquadanio was an assistant professor at the School of Computer Science and Electronic Engineering of the University of Essex, where he received the Wellcome Trust Seed Award in Science to study metabolic switching in renal cell carcinoma.
In 2021, he was awarded the UKRI EPSRC fellowship to design and manufacture enzyme replacement therapies for Fabry’s disease using generative deep learning and yeast. He also collaborates with industrial stakeholders in the biotechnology field, such as Fujifilm Diosynth, on protein design, codon optimisation and lab automation.
Dr Stracquadanio has authored more than 40 research articles published in international peer-reviewed journals, including Science, Nature Rev. Cancer, Cancer Research and PNAS. He also serves as Associate Editor for BMC Genomics and as reviewer and panel member for EPSRC, BBSRC, MRC and FLF. Since 2021, he is also a member of the EPSRC Peer Review Associate College.
.
To join the live event please request the link by emailing: [email protected]
Abstract
“The Landscape of the Heritable Cancer Genome”
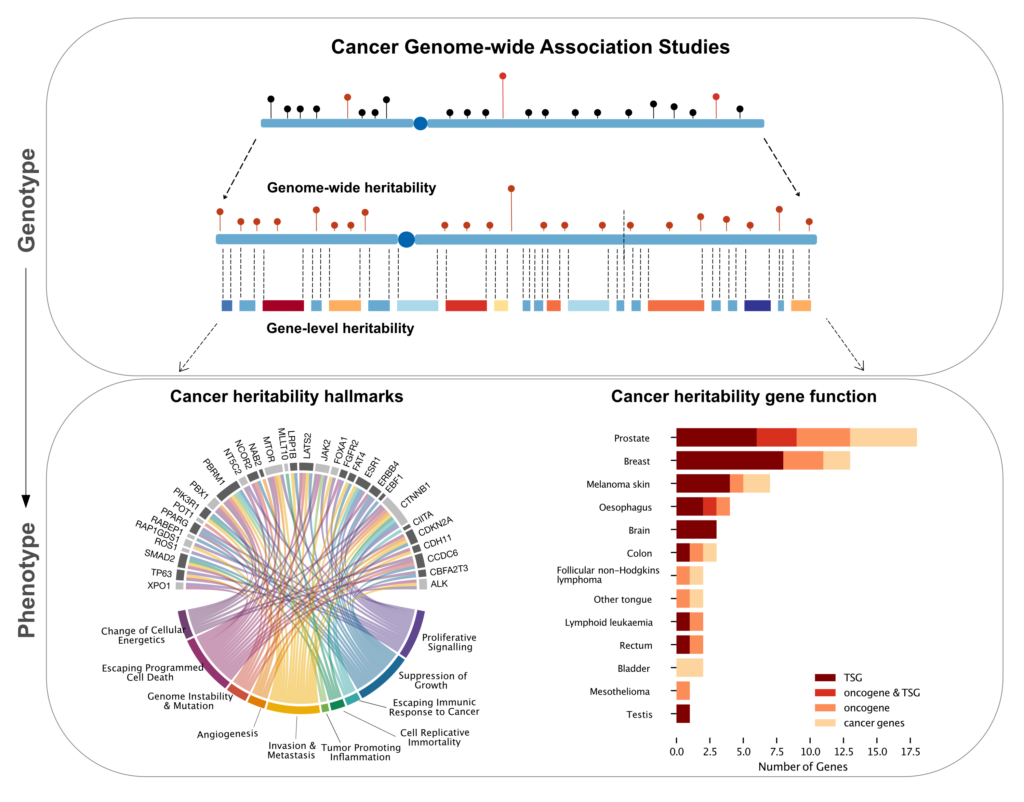
Genome-wide association studies (GWAS) have found hundreds of single nucleotide polymorphisms (SNPs) associated with increased risk of cancer. However, the amount of heritable risk explained by SNPs is limited, leaving most of cancer heritability unexplained. Tumor sequencing projects have shown that causal mutations are enriched in genic regions. We hypothesized that SNPs located in protein coding genes and nearby regulatory regions could explain a significant proportion of the heritable risk of cancer.
To perform gene-level heritability analysis, we developed a new method, called Bayesian Gene HERitability Analysis (BAGHERA), to estimate the heritability explained by all genotyped SNPs and by those located in genic regions using GWAS summary statistics [1]. BAGHERA-based analysis of 38 cancers reported in the UK Biobank showed that SNPs explain at least 10% of the heritable risk for 14 of them, including late onset malignancies. We then identified 1,146 genes, called cancer heritability genes (CHGs), explaining a significant proportion of cancer heritability. CHGs were involved in hallmark processes controlling the transformation from normal to cancerous cells. Importantly, 60 of them also harbored somatic driver mutations, and 27 are tumor suppressors. Evidence for a causal role of CHGs was shown in testicular cancer, where a cluster of SNPs in the KITLG CHG is responsible for p53-mediated responses to genotoxic therapies [2].
Our results suggest that germline and somatic mutation information could be exploited to identify subgroups of individuals at higher risk of cancer in the broader population and could prove useful to establish strategies for early detection and cancer surveillance.